| Product Includes | Mini Kit (P) Quantity | Standard Kit (S) Quantity | Storage Temp |
|---|---|---|---|
| Concanavalin A Magnetic Beads | 1 x 80 µl | 1 x 240 µl | +4°C |
| Concanavalin A Bead Activation Buffer | 1 x 1.7 mL | 1 x 5 mL | +4°C |
| Antibody Binding Buffer (CUT&RUN, CUT&Tag) 15338 | 1 x 800 µl | 1 x 2.5 ml | +4°C |
| 10X Wash Buffer (CUT&RUN, CUT&Tag) 31415 | 1 x 4.5 mL | 1 x 15 ml | +4°C |
| CUT&RUN DNA Extraction Buffer 42015 | 1 x 3.6 mL | 1 x 7 ml | +4°C |
| Calcium Chloride | 1 x 25 µl | 1 x 100 µl | +4°C |
| pAG-MNase Enzyme | 1 x 20 µl | 1 x 40 µl | -20°C |
| Digitonin Solution 16359 | 1 x 1.2 mL | 2 x 1.2 ml | -20°C |
| CUT&RUN 4X Stop Buffer 48105 | 1 x 300 µl | 1 x 1 ml | -20°C |
| 100X Spermidine 27287 | 1 x 400 µl | 1 x 1.3 ml | -20°C |
| Protease Inhibitor Cocktail (200X) 7012 | 1 x 200 µl | 1 x 750 µl | -20°C |
| Proteinase K (20 mg/ml) 10012 | 1 x 20 µl | 1 x 100 µl | -20°C |
| RNAse A (10 mg/ml) 7013 | 1 x 20 µl | 1 x 50 µl | -20°C |
| Tri-Methyl-Histone H3 (Lys4) (C42D8) Rabbit mAb 9751 | 1 x 20 µl | 1 x 20 µl | -20°C |
| Rabbit (DA1E) mAb IgG XP® Isotype Control (CUT&RUN) 66362 | 1 x 20 µl | 1 x 100 µl | -20°C |
| Sample Normalization Spike-In DNA (1 ng/μl) | 1 x 40 µl | 1 x 120 µl | -20°C |
| Sample Normalization Primer Set | 1 x 64 µl | 1 x 150 µl | -20°C |
| SimpleChIP® Human RPL30 Exon 3 Primers 7014 | 1 x 50 µl | 1 x 150 µl | -20°C |
| SimpleChIP® Mouse RPL30 Intron 2 Primers 7015 | 1 x 50 µl | 1 x 150 µl | -20°C |
C&R
Product Information
Storage
Specificity / Sensitivity
Product Description
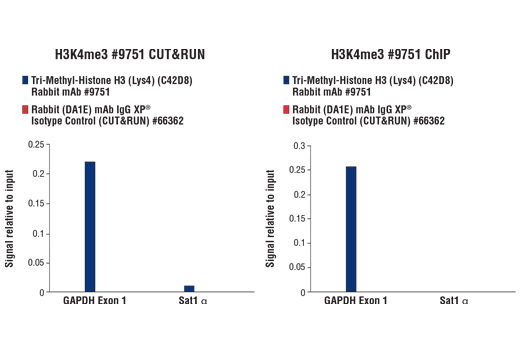
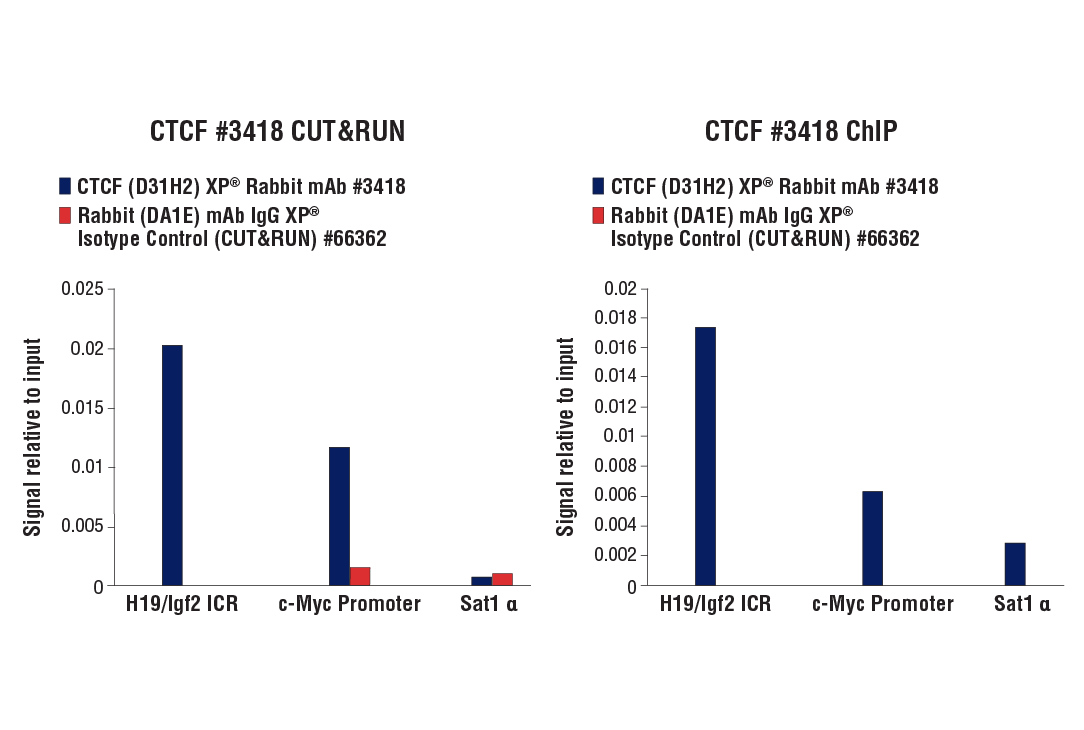
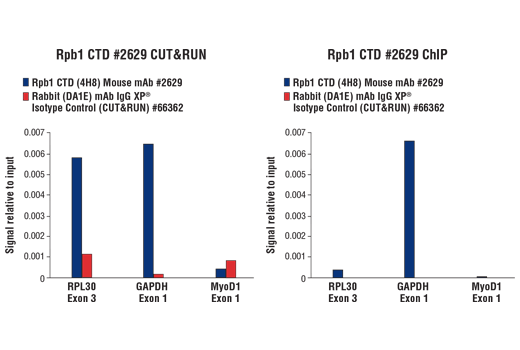
The CUT&RUN Assay Kit also provides important controls to ensure a successful CUT&RUN experiment. The kit contains a positive control Tri-Methyl-Histone H3 (Lys4) (C42D8) Rabbit mAb #9751 and a negative control Rabbit (DA1E) mAb IgG XP® Isotype Control (CUT&RUN) #66362, both of which can be used for qPCR or Next Generation sequencing (NG-seq) analysis. PCR primer sets are provided for the human (#7014) and mouse (#7015) RPL30 gene locus to be used in conjunction with the control antibodies. This kit is compatible with both qPCR and NG-seq.
Background
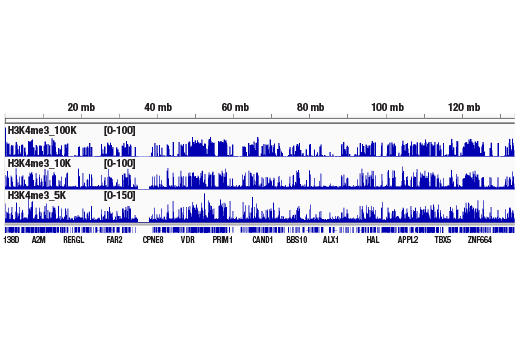
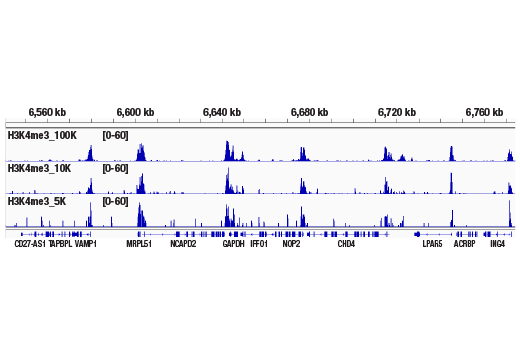
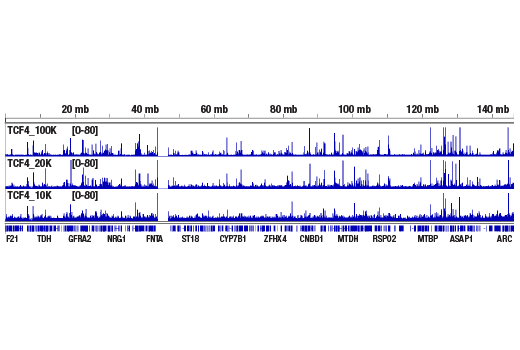
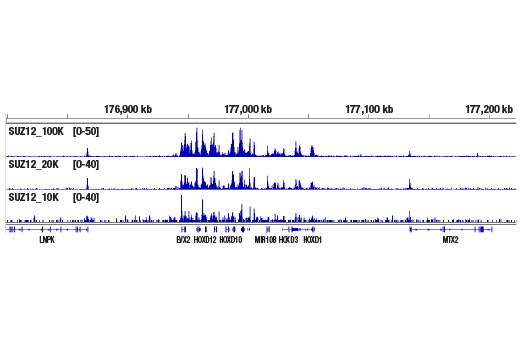
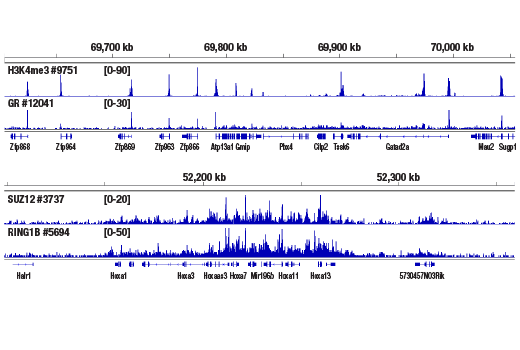
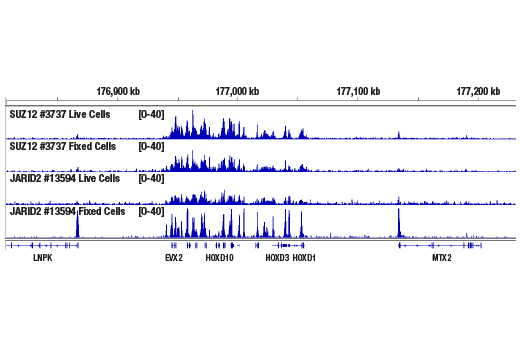
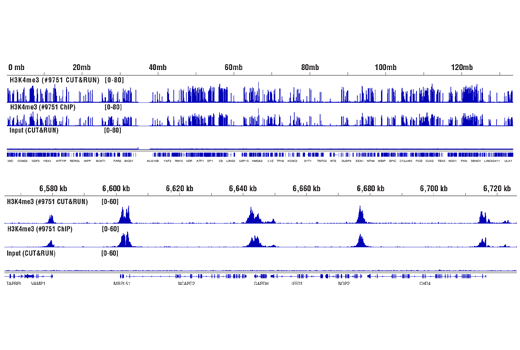
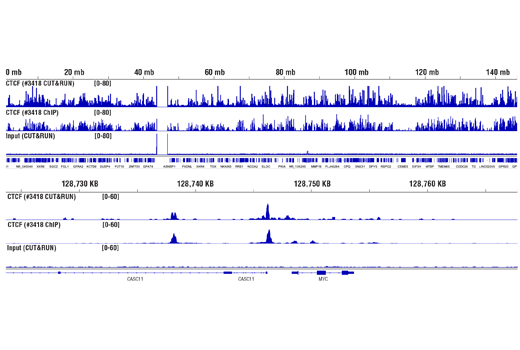
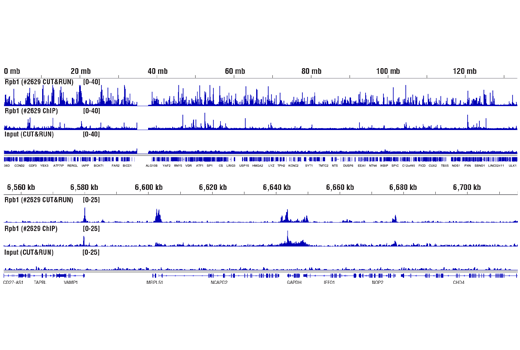
Like the chromatin immunoprecipitation (ChIP) assay, Cleavage Under Targets & Release Using Nuclease (CUT&RUN) is a powerful and versatile technique used for probing protein-DNA interactions within the natural chromatin context of the cell (1-4). This assay can be used to identify multiple proteins associated with a specific region of the genome, or the opposite, to identify the many regions of the genome associated with a particular protein. In addition, the CUT&RUN assay can be used to define the spatial and temporal relationship of a particular protein-DNA interaction. For example, the CUT&RUN assay can be used to determine the specific order of recruitment of various protein factors to a gene promoter or to “measure” the relative amount of a particular histone modification across an entire gene locus during gene activation. In addition to histone proteins, the CUT&RUN assay can also be used to analyze binding of transcription factors and cofactors, DNA replication factors, and DNA repair proteins (Figures 1-6).CUT&RUN provides a rapid, robust, and true low cell number assay for detection of protein-DNA interactions in the cell. Unlike the ChIP assay, CUT&RUN is free from formaldehyde cross-linking, chromatin fragmentation, and immunoprecipitation, making it a much faster and more efficient method for enriching protein-DNA interactions and identifying target genes. CUT&RUN can be performed in less than one day, from live cells to purified DNA, and has been shown to work with as few as 500-1000 cells per assay (1,2). Instead of fragmenting all of the cellular chromatin as done in ChIP, CUT&RUN utilizes an antibody-targeted digestion of chromatin, resulting in much lower background signal than seen in the ChIP assay. As a result, CUT&RUN requires only 1/10th of the sequencing depth that is required for ChIP-seq assays (1,2). Finally, the inclusion of simple spike-in control DNA allows for accurate quantification and normalization of target-protein binding that is not possible with the ChIP method. This provides for effective normalization of signal between samples and between experiments.
Species Reactivity
Species reactivity is determined by testing in at least one approved application (e.g., western blot).
Applications Key
C&R: CUT&RUN
Cross-Reactivity Key
H: human M: mouse R: rat Hm: hamster Mk: monkey Vir: virus Mi: mink C: chicken Dm: D. melanogaster X: Xenopus Z: zebrafish B: bovine Dg: dog Pg: pig Sc: S. cerevisiae Ce: C. elegans Hr: horse GP: Guinea Pig Rab: rabbit All: all species expected
Trademarks and Patents
使用に関する制限
法的な権限を与えられたCSTの担当者が署名した書面によって別途明示的に合意された場合を除き、 CST、その関連会社または代理店が提供する製品には以下の条件が適用されます。お客様が定める条件でここに定められた条件に含まれるものを超えるもの、 または、ここに定められた条件と異なるものは、法的な権限を与えられたCSTの担当者が別途書面にて受諾した場合を除き、拒絶され、 いかなる効力も効果も有しません。
研究専用 (For Research Use Only) またはこれに類似する表示がされた製品は、 いかなる目的についても FDA または外国もしくは国内のその他の規制機関により承認、認可または許可を受けていません。 お客様は製品を診断もしくは治療目的で使用してはならず、また、製品に表示された内容に違反する方法で使用してはなりません。 CST が販売または使用許諾する製品は、エンドユーザーであるお客様に対し、使途を研究および開発のみに限定して提供されるものです。 診断、予防もしくは治療目的で製品を使用することまたは製品を再販売 (単独であるか他の製品等の一部であるかを問いません) もしくはその他の商業的利用の目的で購入することについては、CST から別途許諾を得る必要があります。 お客様は以下の事項を遵守しなければなりません。(a) CST の製品 (単独であるか他の資材と一緒であるかを問いません) を販売、使用許諾、貸与、寄付もしくはその他の態様で第三者に譲渡したり使用させたりしてはなりません。また、商用の製品を製造するために CST の製品を使用してはなりません。(b) 複製、改変、リバースエンジニアリング、逆コンパイル、 分解または他の方法により製品の構造または技術を解明しようとしてはなりません。また、 CST の製品またはサービスと競合する製品またはサービスを開発する目的で CST の製品を使用してはなりません。(c) CST の製品の商標、商号、ロゴ、特許または著作権に関する通知または表示を除去したり改変したりしてはなりません。(d) CST の製品をCST 製品販売条件(CST’s Product Terms of Sale) および該当する書面のみに従って使用しなければなりません。(e) CST の製品に関連してお客様が使用する第三者の製品またはサービスに関する使用許諾条件、 サービス提供条件またはこれに類する合意事項を遵守しなければなりません。