| Product Includes | Product # | Quantity | Color | Storage Temp |
|---|---|---|---|---|
| Spike Protein Coated Microwells II | 87455 | 96 tests |
|
+4C |
| Anti-Human IgM, HRP-linked Antibody (ELISA Formulated) | 15794 | 1 ea |
|
+4C |
| Sample Diluent A | 71637 | 25 ml |
|
+4C |
| HRP Diluent | 13515 | 11 ml |
|
+4C |
| ELISA Wash Buffer (20X) | 9801 | 25 ml |
|
+4C |
| TMB Substrate | 7004 | 11 ml |
|
+4C |
| STOP Solution | 7002 | 11 ml |
|
+4C |
| Sealing Tape | 54503 | 2 ea |
|
+4C |
| ELISA Kit #37322 Positive Control | 60497 | 1 ea |
|
+4C |
| ELISA Kit #37322 Negative Control | 33807 | 1 ea |
|
+4C |
*The microwell plate is supplied as 12 8-well modules - Each module is designed to break apart for 8 tests.
Description
The SARS-CoV-2 Spike Protein Serological IgM ELISA Kit is a solid-phase ELISA that detects binding of human IgM to full-length ectodomain SARS-CoV-2 spike trimeric protein (S-protein). Trimeric spike protein has been coated onto microwells. After incubation with sample, human IgM specific for trimeric spike protein is captured on the plate. The wells are then washed to remove unbound material. Anti-Human IgM, HRP-linked antibody is then used to recognize the bound IgM. HRP substrate, TMB, is added to develop color. The magnitude of optical density for this developed color is proportional to the quantity of IgM specific for spike protein.
*Antibodies in this kit are custom formulations specific to kit.
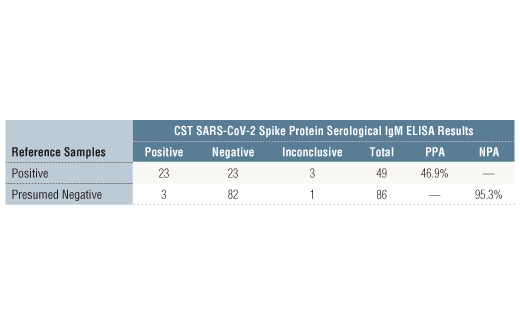
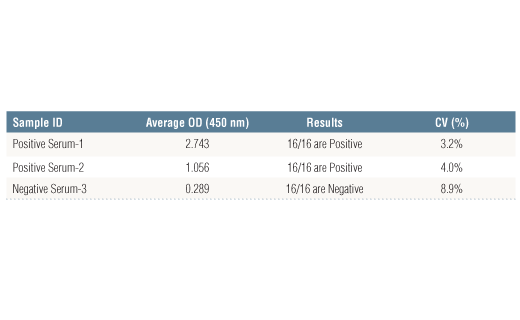
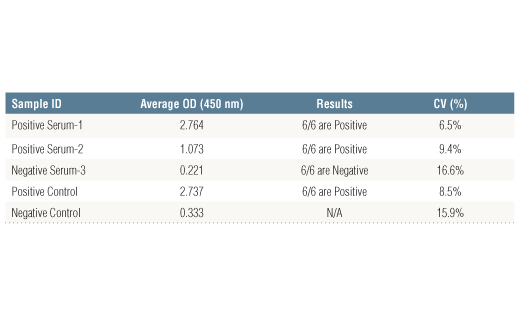
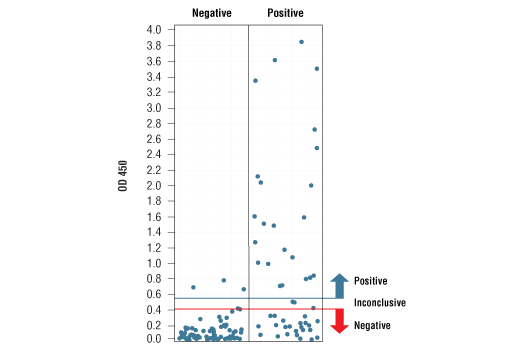
Specificity/Sensitivity
Background
The cause of the COVID-19 pandemic is a novel and highly pathogenic coronavirus, termed SARS-CoV-2 (severe acute respiratory syndrome coronavirus-2). SARS-CoV-2 is a member of the Coronaviridae family of viruses (1). The genome of SARS-CoV-2 is similar to other coronaviruses, and is comprised of four key structural proteins: S, the spike protein, E, the envelope protein, M, the membrane protein, and N, the nucleocapsid protein (2). Coronavirus spike proteins are class I fusion proteins and harbor an ectodomain, a transmembrane domain, and an intracellular tail (3,4). The highly glycosylated ectodomain projects from the viral envelope surface and facilitates attachment and fusion with the host cell plasma membrane. The ectodomain can be further subdivided into host receptor-binding domain (RBD) (S1) and membrane-fusion (S2) subunits, which are produced upon proteolysis by host proteases at S1/S2 and S2’ sites. S1 and S2 subunits remain associated after cleavage and assemble into crown-like homotrimers (2,4). In humans, both SARS-CoV and SARS-CoV-2 spike proteins utilize the angiotensin-converting enzyme 2 (ACE2) protein as a receptor for cellular entry (5-7). Spike protein subunits represent a key antigenic feature of coronavirus virions, and therefore represent an important target of vaccines, novel therapeutic antibodies, and small-molecule inhibitors (8,9).
- Zhou, P. et al. (2020) Nature 579, 270-3.
- Tortorici, M.A. and Veesler, D. (2019) Adv Virus Res 105, 93-116.
- Li, F. et al. (2006) J Virol 80, 6794-800.
- Li, F. (2016) Annu Rev Virol 3, 237-61.
- Shang, J. et al. (2020) Nature 581, 221-4.
- Wrapp, D. et al. (2020) Science 367, 1260-3.
- Yan, R. et al. (2020) Science 367, 1444-8.
- Yuan, Y. et al. (2017) Nat Commun 8, 15092.
- Amanat, F. and Krammer, F. (2020) Immunity 52, 583-9.
Background References
Cross-Reactivity Key
H: human M: mouse R: rat Hm: hamster Mk: monkey Vir: virus Mi: mink C: chicken Dm: D. melanogaster X: Xenopus Z: zebrafish B: bovine Dg: dog Pg: pig Sc: S. cerevisiae Ce: C. elegans Hr: horse GP: Guinea Pig Rab: rabbit All: all species expected
Trademarks and Patents
使用に関する制限
法的な権限を与えられたCSTの担当者が署名した書面によって別途明示的に合意された場合を除き、 CST、その関連会社または代理店が提供する製品には以下の条件が適用されます。お客様が定める条件でここに定められた条件に含まれるものを超えるもの、 または、ここに定められた条件と異なるものは、法的な権限を与えられたCSTの担当者が別途書面にて受諾した場合を除き、拒絶され、 いかなる効力も効果も有しません。
研究専用 (For Research Use Only) またはこれに類似する表示がされた製品は、 いかなる目的についても FDA または外国もしくは国内のその他の規制機関により承認、認可または許可を受けていません。 お客様は製品を診断もしくは治療目的で使用してはならず、また、製品に表示された内容に違反する方法で使用してはなりません。 CST が販売または使用許諾する製品は、エンドユーザーであるお客様に対し、使途を研究および開発のみに限定して提供されるものです。 診断、予防もしくは治療目的で製品を使用することまたは製品を再販売 (単独であるか他の製品等の一部であるかを問いません) もしくはその他の商業的利用の目的で購入することについては、CST から別途許諾を得る必要があります。 お客様は以下の事項を遵守しなければなりません。(a) CST の製品 (単独であるか他の資材と一緒であるかを問いません) を販売、使用許諾、貸与、寄付もしくはその他の態様で第三者に譲渡したり使用させたりしてはなりません。また、商用の製品を製造するために CST の製品を使用してはなりません。(b) 複製、改変、リバースエンジニアリング、逆コンパイル、 分解または他の方法により製品の構造または技術を解明しようとしてはなりません。また、 CST の製品またはサービスと競合する製品またはサービスを開発する目的で CST の製品を使用してはなりません。(c) CST の製品の商標、商号、ロゴ、特許または著作権に関する通知または表示を除去したり改変したりしてはなりません。(d) CST の製品をCST 製品販売条件(CST’s Product Terms of Sale) および該当する書面のみに従って使用しなければなりません。(e) CST の製品に関連してお客様が使用する第三者の製品またはサービスに関する使用許諾条件、 サービス提供条件またはこれに類する合意事項を遵守しなければなりません。